-
Notifications
You must be signed in to change notification settings - Fork 674
Description
This issue references the following line of code:
The documentation accurately describes the standard_scale='var' normalization strategy as
"Whether or not to standardize the given dimension between 0 and 1, meaning for each variable or group, subtract the minimum and divide each by its maximum."
Something about this normalization has bothered me for a long time, and I finally realized: it's the subtraction of the minimum value. This subtraction means that the minimum valued dot will have color = 0. Imagine a case with only two groupby groupings: healthy and disease. In that case, one of the dots will always have color 0, and the other will have color 1. Totally binary, no matter how close the actual values are.
I feel that this kind of normalization is very misleading for scRNA-seq data!
A random example follows:
This image makes it look like these genes are very specific to one tissue or another ---------------
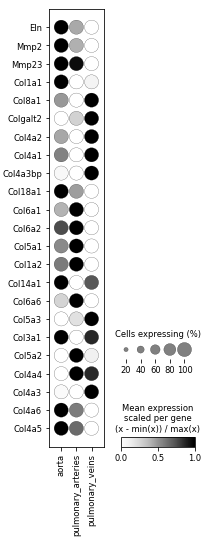
But in reality, if we had scaled by just dividing by the max (and not first subtracting the min), then we'd see -------
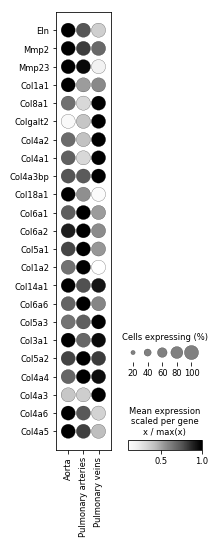
which is much more realistic, and much closer to what you'd see if you used a log-scaling instead, and made several different plots for genes expressed at different levels overall.
I leave it up for discussion, but I would suggest two types of fixes:
- delete the above line 185 (and the other places it shows up...)
- allow the user to specify a custom normalization function (but change the default to
x / max(x)instead of(x - min(x)) / max(x))