Venue: MIDL 2025
Paper: Conditional Diffusion Models are Medical Image Classifiers that Provide Explainability and Uncertainty for Free
Authors: Gian Favero*, Parham Saremi*, Emily Kaczmarek, Brennan Nichyporuk, Tal Arbel
Institution(s): Mila - Quebec AI Institute, McGill University
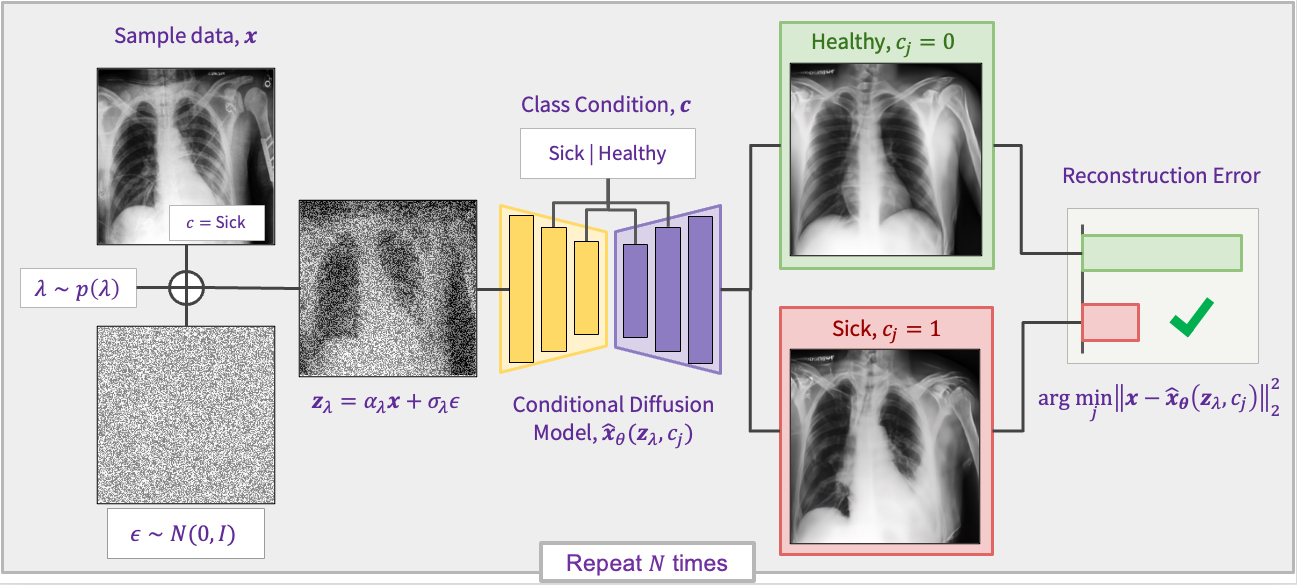
- Use Linux (recommended) for best performance, compatibility, and reproducibility.
- All testing, training, inference completed with A100 NVIDIA GPUs (single or multiple).
- 64-bit Python 3.10 and PyTorch 2.6. See https://pytorch.org for PyTorch install instructions.
- Python virtual environment (recommended) to manage libraries, packages for this repository.
First, clone this repository.
Required packages are provided in the requirements.txt file and can be installed using the following command:
pip install -r requirements.txtFlashAttention can be installed for faster inference time (especially for DiT models). The wheel file can be downloaded from FlashAttention GitHub. We used flash_attn-2.7.4.post1+cu12torch2.6cxx11abiFALSE-cp310-cp310-linux_x86_64.whl for our CUDA and Torch version. After downloading the wheel file, install it with:
wget https://github.com/Dao-AILab/flash-attention/releases/download/v2.7.4.post1/flash_attn-2.7.4.post1+cu12torch2.6cxx11abiFALSE-cp310-cp310-linux_x86_64.whl
pip install flash_attn-2.7.4.post1+cu12torch2.6cxx11abiFALSE-cp310-cp310-linux_x86_64.whlWe use the CheXpert and ISIC datasets in our paper. Our train/validation/test CSV files for both datasets are in the splits folder.
All trained models can be downloaded from this link. Alternatively, use the following command with gdown:
gdown --folder 1x7CKrbS8pxS45EXzpUKhpusBYCLgH82YBefore using the models, modify the scripts/run.sh file:
PROJECT_ROOT: Absolute path to the root directory of the diffusion-classifier repository.DATA_ROOT: Absolute path to the data directory containingisic_mel_balanced/,chexpert/, andsd_isic_chexpert/folders.INFERENCE_CHECKPOINT_FOLDER: Absolute path to the directory where the downloaded model weights are stored.
If you want to use CometML for experiment tracking, set the COMET variables in run.sh. Additionally, you must set USE_COMET=1 to enable tracking.
All training/inference hyperparameters are defined in their corresponding bash scripts. For example, the CheXpert-UNet hyperparameters are specified in scripts/unet/chexpert-unet.sh.
Below, we describe various use cases that are easily achievable with simple customizations to our code. In any case, launching the desired experiment is done via bash scripts/run.sh from the parent folder of the repository.
Scripts to run inference with all models are provided in the scripts folder. However to launch each script you only have to modify the run.sh file to select which model and data you want to run. For instance, to run the UNet model's inference on the CheXpert dataset, you'll need to set MODEL=unet, DATA=chexpert, FUNCTION=inference.
Baseline classifiers for both datasets can be evaluated using scripts in scripts/run.sh. You can change the VARIANT and BACKBONE environment variables to run different models. Available models:
| VARIANT | BACKBONE |
|---|---|
| resnet18 | resnet |
| resnet50 | resnet |
| efficientnet_b0 | efficientnet |
| efficientnet_b4 | efficientnet |
| swin_base_patch4_window7_224 | swin |
| vit_base_patch16_224 | vit |
| vit_small_patch16_224 | vit |
To evaluate on CheXpert, modify the run.sh file:
export MODEL="baseline"
export FUNCTION="inference"
export DATA="chexpert" Or on ISIC:
export MODEL="baseline"
export FUNCTION="inference"
export DATA="chexpert" Use the following instructions to run inference with diffusion models.
For faster inference, set the FLASH_ATTENTION variable to true for DiT and UNet models.
CheXpert-UNet:
export MODEL="unet"
export FUNCTION="inference"
export DATA="chexpert" ISIC-UNet:
export MODEL="unet"
export FUNCTION="inference"
export DATA="isic" CheXpert-DiT:
export MODEL="dit"
export FUNCTION="inference"
export DATA="chexpert" ISIC-DiT:
export MODEL="dit"
export FUNCTION="inference"
export DATA="isic" CheXpert-StableDiffusion:
export MODEL="sd"
export FUNCTION="inference"
export DATA="chexpert" ISIC-StableDiffusion:
export MODEL="sd"
export FUNCTION="inference"
export DATA="dit" Counterfactual generation is currently supported only for UNet models. To generate counterfactuals for the UNet models, modify the script to run explain.py instead of inference.py. This can be easily done by changing the FUNCTION value to explain:
export MODEL="unet"
export FUNCTION="explain"
export DATA="chexpert" To improve visual quality, increase SAMPLING_STEPS to at least 256 in the unet scripts (scripts/unet/chexpert-unet.sh and scripts/unet/isic-unet.sh). CFG_W refers to the classifier-free guidance scale.
The images will be saved in the inference_images directory located within the experiment folder.
Similar to counterfactual generation, training different models is as simple as changing the FUNCTION to train. For example, to train the UNet model on CheXpert data you should use the following environment variables:
export MODEL="unet"
export FUNCTION="train"
export DATA="chexpert"
Note: The Stable Diffusion model is jointly trained on both ISIC and CheXpert datasets. To train on a single dataset, modify the metadata.jsonl placed in data/sd_isic_chexpert file and adjust the training data folder accordingly.
In order to train the Stable Diffusion model, the diffusers package should be installed from source. To do this you can run the following command:
pip install git+https://github.com/huggingface/diffusers
The output directory for the Stable Diffusion model can be set in its train.sh script, while other models will save their checkpoints within their respective experiment folders.
This project is licensed under the MIT License. See the LICENSE file for details.
@misc{favero2025conditionaldiffusionmodelsmedical,
title={Conditional Diffusion Models are Medical Image Classifiers that Provide Explainability and Uncertainty for Free},
author={Gian Mario Favero and Parham Saremi and Emily Kaczmarek and Brennan Nichyporuk and Tal Arbel},
year={2025},
eprint={2502.03687},
archivePrefix={arXiv},
primaryClass={cs.CV},
url={https://arxiv.org/abs/2502.03687},
}

