Attention Attention Attention
We are currently rewriting Farseer-NMR towards version 2.
Version 1 is still functional and working, though not much supported apart from minor bugs. You can download the latest stable version, v1.3.5, on the releases tab, or visit the complete version 1 code and its documentation in version 1 branch) on GitHub.
Our original publication is available at JBioMolNMR), cite us if you use Farseer-NMR for your research, regardless of which version you use.
Please note that the master branch) currently hosts the development of version 2, which is UNFINISHED software; again, please, refer to version 1 for a stable and functional release.
Farseer-NMR runs purely on volunteer work without any official assigned funds. All help is welcomed, engage with us)!
Attention Attention Attention
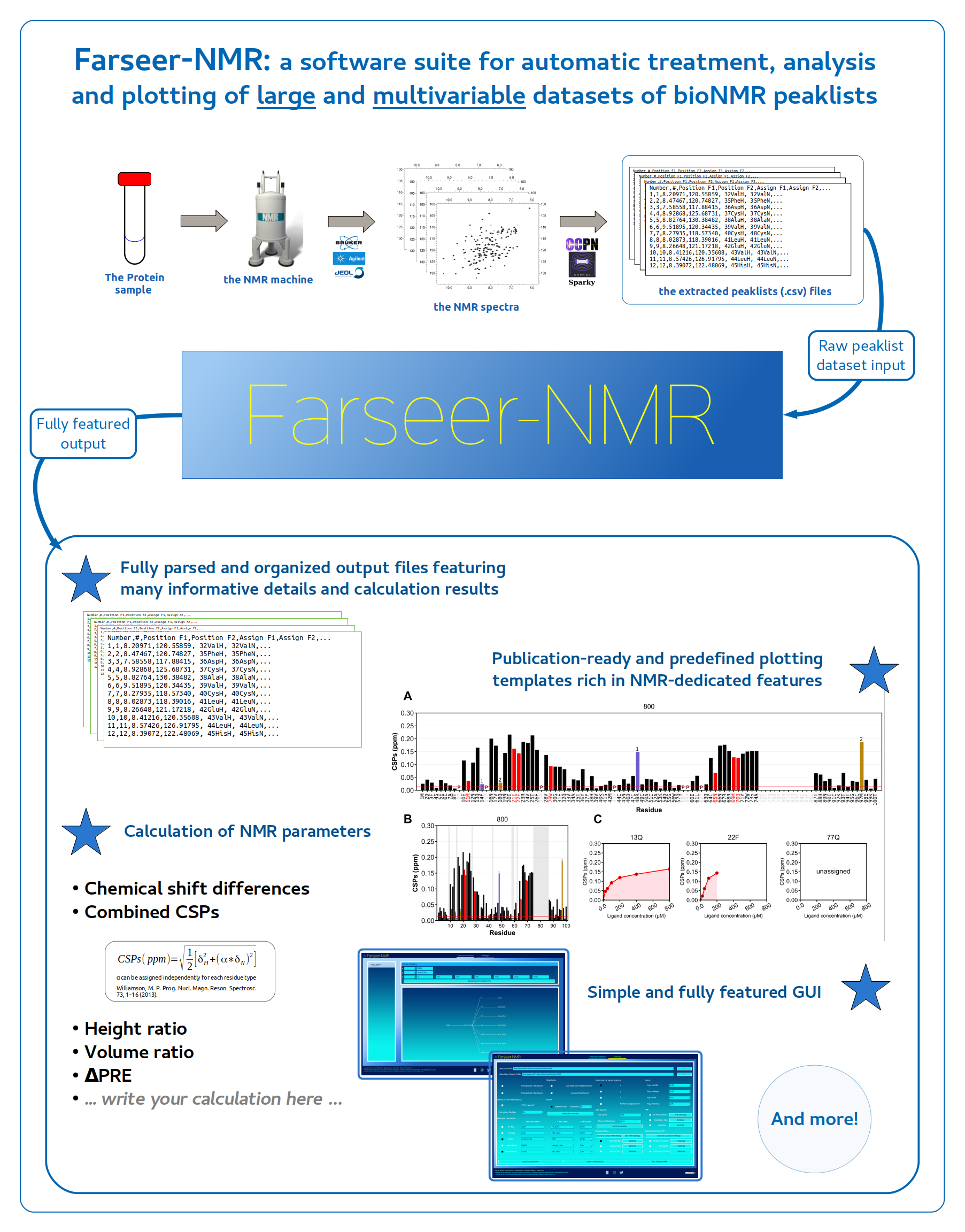
A Python written, multi-platform and fully community-driven suite to analyse datasets of peaklist files extracted from multivariable series of Biomolecular Nuclear Magnetic Resonance (NMR) experiments.
With Farseer-NMR, you have:
- Automatic analysis of large and multivariable NMR peaklist files datasets
- Peaklist parsing and treatment
- Identification of _missing_ and _unassigned_ residues
- Automatic calculation of NMR parameters
- Comprehensive organization of the output
- Large suite of publication-ready plotting templates
- Full traceability via Markdown) formatted log file.
Download here the latest version of Farseer-NMR..
To install Farseer-NMR simply run the installation script:
python install_farseernmr.py
Read here some additional detail on how to setup your Farseer-NMR installation - it's very easy!
The complete Farseer-NMR documentation is available on our Wiki page.
To contribute to the development of Farseer-NMR visit our GitHub project). If you are a user, share your experience in the issues tab (reporting bugs, suggestions or discussions); if you are a developer read our CONTRIBUTING) guidelines.
- Post on our mailing list) for questions, discussion and help!
- Follow us on Twitter @farseer_nmr)
- Find us on Research Gate), where our seminars PDFs are available!
Thanks for using Farseer-NMR!
If you are using Farseer-NMR, or any of its components, to analyze your NMR peaklist data, please cite our original article:
Teixeira, J.M.C., Skinner, S.P., Arbesú, M., Breeze, A.L., Pons, M. J Biomol NMR (2018) 71:1, 1-9. DOI 10.1007/s10858-018-0182-5)
- Miguel Arbesú, MiquelPons. Integrating disorder in globular multidomain proteins: Fuzzy sensors and the role of SH3 domains. Archives of Biochemistry and Biophysics 2019, 677, 108161 https://doi.org/10.1016/j.abb.2019.108161.
- Luca Mureddu, Geerten W. Vuister. Simple high‐resolution NMR spectroscopy as a tool in molecular biology. The FEBS Journal 2019, 286, issue 11, p2035 https://doi.org/10.1111/febs.14771.
- Teixeira, J.M.C.; Fuentes, H.; Bielskutė, S.; Gairi, M.; Żerko, S.; Koźmiński, W.; Pons, M. The Two Isoforms of Lyn Display Different Intramolecular Fuzzy Complexes with the SH3 Domain. Molecules 2018, 23, 2731. https://doi.org/10.3390/molecules23112731)
- Arbesú, M.; Iruela, G.; Fuentes, H.; Teixeira, J.M.C.; Pons, M. Intramolecular fuzzy interactions involving intrinsically disordered domains. Front. Mol. Biosci. 2018, 5, 39. DOI 10.3389/fmolb.2018.00039)
- Arbesú, M. et al. (2017) The Unique Domain Forms a Fuzzy Intramolecular Complex in Src Family Kinases. Structure 25, 630–640.e4. 10.1016/j.str.2017.02.011)
- Marimon, O. et. al. (2016). An oxygen-sensitive toxin–antitoxin system. Nature Communications, 7, 13634. https://doi.org/10.1038/ncomms13634)
- Bijlmakers, M.-J., et.al. (2015) A C2HC zinc finger is essential for the RING-E2 interaction of the ubiquitin ligase RNF125. Scientific Reports, 6, 29232. https://doi.org/10.1038/srep29232)
Farseer-NMR on Google Citations) .. end-citing .. start-acknowledge
The Farseer-NMR Project wants to acknowledge to following people for their contributions to the project:
- Susana Barrera-Vilarmau (ORCID 0000-0003-4868-6593): beta-tester, data provider, plot suggestions.
- Jamie Ferrar, Artistic Systems: for providing the UI branding.
- João P.G.L.M. Rodrigues: for all the years of coding discussions and mentorship, and in particular for the help in setting the Farseer-NMR organization profile on GitHub.
- Héctor Fuentes: intensive beta-tester, specially for the Windows version.
- To all the users and participants of the Farseer-NMR workshops, thanks for your feedback, opinions, testing, interest and patience. Your contribution is definitively making Farseer-NMR growing bigger and robust! Special thanks to:
The entire Farseer-NMR project is distributed with no liability and is licensed under the GPL-3.0.
<a href="https://www.gnu.org/licenses/gpl-3.0.en.html"><img src="https://upload.wikimedia.org/wikipedia/commons/thumb/9/93/GPLv3_Logo.svg/1200px-GPLv3_Logo.svg.png" width="75" height="37"></a>